Machine Learning Project Workflow - part I
TL;DR
This is the first part of a blog post series about machine learning workflow. The first part focuses on the code management in machine learning projects. Overall, this blog post series will try to give valuable advice on:
- how to bring peace and order into code and data chaos created by many experiments in machine learning project
- how to easily share our experiments with colleagues
- how to manage data sets
- how to easily visualize the results of experiments
- how to utilize cloud power in machine learning projects
- how to automate the whole machine learning workflow
- how to efficiently put machine learning models into production
Introduction
At the beginning of every machine learning journey there is a problem that needs a solution. The solution is a machine learning model. To build a model, you need the ingredients (data) and a recipe (code). In a few minutes, hours or sometimes days you get your model and the problem seems to be solved. Easy, right?

It is a super rare case when the first experiment aka baseline experiment outputs the model with satisfying performance based on the defined metrics (confusion matrix, accuracy, precision, recall, f1 score or others). Often, one needs to change the preprocessing or tune model parameters (number of hidden layers, activation function and so on) or training parameters (optimization function, learning rate and parameters) to create a desired model with acceptable performance. When this happens, you start modifying your recipe (code) and thus creating your second, third, fourth or n-th experiment with modified parameters.
And now is the time when everyone that takes machine learning seriously has to ask several questions, mainly
- When I run multiple experiments, how can I efficiently manage data files so that common data files for each experiment are easily sharable across experiments?
- How to easily redefine and rerun ML pipeline with as little steps as possible?
- How to store results of every experiment so that they are easily accessible and comparable among various experiments?
- If my colleague wants to have a look at the experiments and possibly revisit them how can I easily share with her/him the whole code, environment setup and data?
If you continue creating and running experiments without answering questions like these then at some point, the whole project will be a mess and full of chaos where no one knows which experiments have been tried so far and the experiments will have to be rerun once again. The project will be also hardly shareable with other people.
The result of not answering the questions above is shown in the picture. This is how it ends up … in total chaos.
As a result of not answering these questions beforehand you can get from this
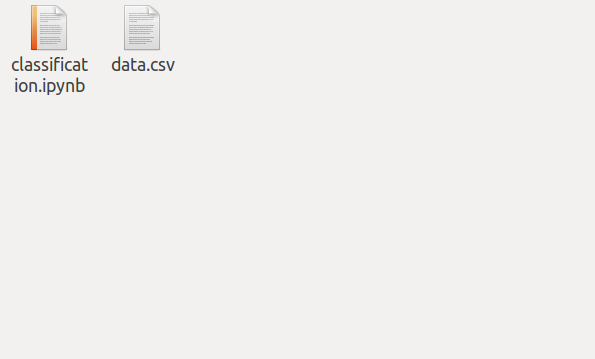
to this mess
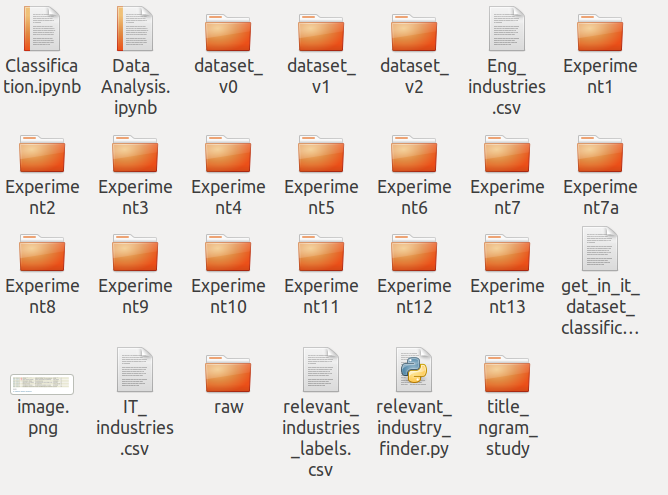
This also happened to us and we had to do a bit of research on how to manage the machine learning experiments. So let’s have a look at how we, at Experteer, tackled this challenge when we started taking machine learning projects seriously.
In the first part of this blog post series we will answer only a few of aforementioned questions. It will be mainly the ones involving options to write a cleaner, shareable and readable code for machine learning projects while considering all implications from answering the other aforementioned questions. I will also present you the way that we think is the best.
Code Management
In machine learning project, code management breaks down into two major planes i.e. how/where to write machine learning recipe(code) and how to manage code for various experiments efficiently.
How/where to write the code in machine learning project
In machine learning or data science, there are two ways how to write code.
The first one is Jupyter Notebook.
Jupyter Notebook is an open-source web application that offers users to create documents with code, equations, visualizations and narrative text.
Nowadays, there is also JupyterLab which offers whole interactive development environment for Jupyter notebooks.
You can find more information here.
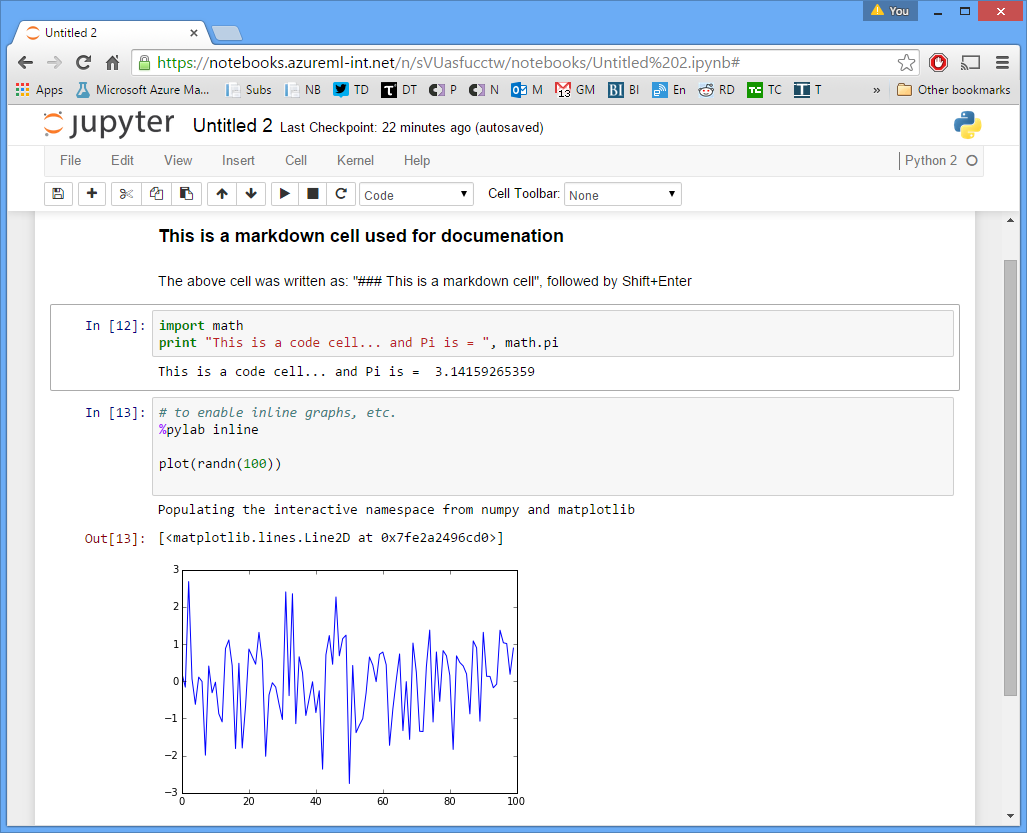 Jupyter Notebook is structured into cells.
Once a cell is run the state of the cell is preserved and accessible to other cells until it is rerun again.
Jupyter Notebook is structured into cells.
Once a cell is run the state of the cell is preserved and accessible to other cells until it is rerun again.
Due to the nature of Jupyter Notebook almost every beginner in machine learning or any data related field starts writing the code in Jupyter Notebook because, as opposed to regular Python scripts, it allows for quick prototyping and thus speeding up the coding. Since Jupyter Notebook cell preserves the state of the cell, one does not have to rerun the whole notebook when one cell is modified but only the modified cell. This has a big advantage in preprocessing or data analysis because you can separate data set loading into one cell and preprocessing or analytics into another cell. By doing so, when you change something in analytics or preprocessing cells there is no need to rerun the cell that loads data set because it is already in the memory so you only need to rerun the cell where you did the change. And this can save tremendous amount of time when working with a big data set.
On the other hand, as mentioned, Jupyter Notebook is a web application and might cause some troubles. Imagine a situation where you want to run training on a big data set which will take a few hours. Since it is going to take quite a lot of time, you want to track intermediate results such as accuracy, loss, F1 score and so on epoch by epoch by writing it into the cell’s output. Everything works well until you realize that you need to go home and turn off your machine or you accidentally close your notebook. Currently, if you close Jupyter Notebook, your current session will get lost and there is no way to reconnect to it again. You can read more about it here. Jupyter Notebook does not persist sessions so this means that you cannot properly continue working with the training results once you get disconnected from the notebook that started the training. This is the biggest disadvantage of Jupyter Notebook. Although there are some workarounds how to tackle it like the ones mentioned here or here. However, none of them work very well. The most promising I see nowadays is this initiative. Until it gets implemented and released I found the best way to overcome this issue by starting Jupyter server on a remote machine, booting up a virtual machine on the same remote machine and connecting to Jupyter server via the browser from within the virtual machine on the remote machine. Then, the Jupyter Notebook runs on another machine so even turning off your local machine will not cause Jupyter Notebook to disconnect so you do not have to stay at work the whole night and can take your notebook home :) Even though this workaround works, it is still kind of overhead.
The other possibility how/where to write machine learning code is to write it as a regular Python script. This is not that popular since it does not offer quick prototyping as opposed to Jupyter Notebook and it also does not offer an option to divide the code into cells caching the cell state so the whole code must be rerun when it gets modified. Usually, there is no need to work with the whole data set when designing experiment pipeline. In this case, even working with 10 samples is enough and loading speed should be negligible. This way, rerunning the script should be very fast.
After experiments with both approaches, we finally decided to use Python scripts for writing our machine learning recipe (code). However, we also did not get rid of Jupyter notebooks completely. We still use them for analytics and testing our models because this is the field where they can shine the most. The reasons for choosing Python scripts over Jupyter notebooks were mainly:
- Python scripts are easy to share via version controlling system (read more below)
- they better integrate with data versioning system tool we are using (see the second part of this series)
- they are readable when you do git diff
- they do not require open browser to run; they can be started in separate terminal screen so we can easily disconnect from their execution
- and others
How to manage code for various experiments efficiently
Often, machine learning project requires to run more than just one experiment. Usually, experiments follow each other because we want to try different training parameters, algorithms, pre-trained embeddings and models, and so on. One way is to create a separate directory for each experiment but then we will end up with a mess and chaos that we have seen in the second picture and that is not what we want. The other way is to keep it clean as in the first picture but with more experiments in one directory. We can achieve this by overwriting experiments with new experiments but then old experiments get deleted. This approach is kind of silly because new experiment with a new set of model parameters might perform worse than the previous experiment but we already overwrote the previous experiment with the whole recipe (code base) together with all its outputs. Now, the only way to get it back is to rerun the previous experiment which costs time and money. But, are these really the only ways how we can manage the machine learning recipe (code base) for different experiments?
In software engineering projects, developers use version controlling systems like git to keep track of all the changes and share the code easily. Version controlling system can be also leveraged in machine learning projects so we can easily not only share the code and keep track of all the changes but also manage recipe (code) for various experiments.
In software engineering, branches are mainly leveraged to add a piece of additional functionality aka feature. In machine learning, there are no features in this sense but we can leverage branches to manage our experiments instead by assigning each experiment to its own branch. By doing so, we bring order into the project structure and the second picture you have seen above will reduce only to the code that is relevant for the current experiment. Leveraging branches can speed up new experiment creation because we do not have to copy the whole code base to create it. We only create a new branch out of the experiment branch that we want to modify. In other words, we are forking one experiment from another one.
As in software engineering, branch naming follows certain conventions. The same should be applied in machine learning projects. In software engineering, the branch name is usually the name of a feature so the name characterizes the feature that is being added to the software. Since each experiment extends or modifies the previous experiment (unless it is the baseline) I decided to assign each experiment branch a name that characterizes a particular experiment so that it can also serve as a title for the experiment. Recently, we had a project where we wanted to perform NER (named entity recognition). A few experiments tried to leverage Facebook laser encoder in NER model. The experiment branches were named as follows:
- extraction_laser_embeddings (baseline)
- extraction_laser_embeddings_lr_schedular
- extraction_laser_embeddings_lr_schedular_wd_1e-7
- and so on
Summary
In this first part of the machine learning workflow series, I compared different approaches how to take care about machine learning recipe (code base). I also showed you what we, at Experteer, found to be the best way to work with the machine learning recipe. Everything has its pros and cons but for us writing machine learning pipeline code in regular Python scripts, using Jupyter Notebook only for data analytics and testing and version experiments via branches in git is the best way of working on machine learning projects considering also other aspects like data files management and others. The next part on managing data files in machine learning project will show how to succeed in it. It will also prove our decisions made in this part to be the right ones.
